Nottingham FP Lab Blog
braincurry
by Tom Nielsen on July 3, 2008.
Tagged as: Lunches.
Last Friday, I presented the braincurry project, which is my attempt, with assistance from Henrik Nilsson, to create a domain specific language to define and execute experiments and simulations related to cellular neuroscience. I spent a bit of time introducing neurons, dendrites, axons, synapses and the problem of synaptic integration (given a pattern of inputs to a neuron, can we predict its outputs?) I also talked briefly about the preparation I am currently working on in Tom Matheson’s lab in Leicester, the locust visual system and the influence of locust swarming on agriculture and crop failure.
Experiments with invertebrates and in particular insects can include an large number of modalities, including visual, auditory and olfactory sensory stimulation, intracellular or extracellular recording from multiple neurons, and movement observation – all at the same time. I presented a prototype language for defining these experiments and their associated analyses.
I restrict myself to experiments that consists of trials, i.e. finite episodes during which a set of stimuli (visual, waveforms of currents to be injected into neurons, etc) are presented and responses (neuron spikes, etc) are recorded. Analog input and output is controlled from a simple FFI binding to the Comedi library, which in turns controls a National Instruments data acquisition board. All aspects of the stimuli must be known before the beginning of the trial, so there are no dynamic or reactive components, although stimuli can be influenced by the outcome of previous trials. This restriction suggests a very simple definition of a trial, which is a list of trial specifications:
![%format dots = “\ldots”
> type Trial = [TrialSpecification]
> data TrialSpecification = RecordWaveform Channel
> | PlayVideo dots
> | PlayAudio dots
%format dots = “\ldots” > type Trial = [TrialSpecification] > data TrialSpecification = RecordWaveform Channel > | PlayVideo dots > | PlayAudio dots](../images/latexrender/pictures/8ae7a60c1d88d631956fa7c24c84c463.png)
%format dots = “\ldots” > type Trial = [TrialSpecification] > data TrialSpecification = RecordWaveform Channel > | PlayVideo dots > | PlayAudio dots
In addition, I would like to be able to specify analyses that are performed online, ie immediately after a trial has concluded. I can implement this in an untyped manner by giving names to recordings and additionally specify names as inputs and outputs of analyses. First I define a results data type:
![%format dots = “\ldots”
> data Result = ResWaveform [Double]
> | ResNumber Double
> | ResBool Bool | dots
> type Name = String
%format dots = “\ldots” > data Result = ResWaveform [Double] > | ResNumber Double > | ResBool Bool | dots > type Name = String](../images/latexrender/pictures/1f32916b831beb202b06d71fcf526efd.png)
%format dots = “\ldots” > data Result = ResWaveform [Double] > | ResNumber Double > | ResBool Bool | dots > type Name = String
And modify TrialSpecification
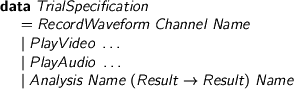
%format dots = “\ldots” > data TrialSpecification > = RecordWaveform Channel Name > | PlayVideo dots > | PlayAudio dots > | Analysis Name (Result -> Result) Name
One advantage of this approach is that I do not have to specify the order in which analyses are carried out if they depend on one another; their dependencies can be resolved automatically, although not in a type-safe manner. Trials are also very easy to compose. We discussed various possibilities for defining a type-safe trial, including monads and GADTs. I talked a bit about how this language may allow me to use parametrised trials to define general higher order trials that can determine, for instance, whether any one trial parameter influences some experimental outcome.
comments powered by Disqus